Seminar Abstract
Bishnubrata Patra, Manvendra Sharma, William Hale, Marcel Utz
University of Southampton
MicroTAS 2018
ABSTRACT
NMR metabolomics, the study of cellular metabolism, provides a new path to understand cellular response in vitro. In this article, we performed a real-time monitoring of adherent vs. spheroid cell culture using NMR metabolomics. The difference in lactate production provides a clear indication of different metabolic rates in spheroid compared to adherent cell culture. The developed microfluidic–NMR technique could be applicable towards real-time monitoring of a variety of ex-vivo biological samples, including tissue or organ on a chip.
KEYWORDS: Cell culture, Spheroids, Microfluidics, NMR
INTRODUCTION
Three-dimensional cell culture models (spheroids) provide valuable additional insight into the behavior of cancer cells since it mimics the features such as cell-cell contact and hindrance of metabolic diffusion which are not present in traditional cell culture.1On the other hand, NMR spectroscopy can provide in-situ monitoring of metabolomics even at microliter volume.2In this work, we used a novel integrated transmission-line NMR probe to directly compare the metabolomic activity of MCF-7 cells in adherent (2D) and spheroid (3D) culture. Microfluidic cell culture device of volume 2.5 μl volume was coated with fibronectin and with pluronic respectively to produce 2D and 3D cell cultures. Metabolomics observation was done up to 48 hours using NMR spectroscopy, followed by viability analysis. Clear differences in metabolomic activity were found, the spheroid (3D) showed much lower consumption of glucose, production of lactate, and acidosis compared to the attached (2D) culture.
EXPERIMENTAL
Microfluidic devices were made from thermally bonded PMMA layers. The cell culture chamber was coated overnight with fibronectin (1 mg/ ml) orpluronic (10 mg/ml). A suspension of 2.5 μl MCF-7 Cells (1000 cells/μl) in DMEM culture medium was introduced into each device. The devices were kept inside a cell culture incubator at 37° C and with 5% CO2. 1H NMR spectra were recorded using a 600 MHz spectrometerforboth2D and 3D cell culture devices and controls (without cells) periodically at 4, 8, 24, 32, and 48 hours. Cell viability was analyzed on the same devices using Live/Dead cell imaging kit [life technologies]and fluorescence microscopy.
RESULTS AND DISCUSSION
In this work, we used the equal number of cells with the equal volume of culture medium and the same culture environment to compare the metabolic activity of 2D vs.3D cultured MCF-7 cells by NMR spectroscopy. Our results showed that aggregated cells are metabolically less active and more viable after 48 hours compared to the attached cells
CONCLUSION
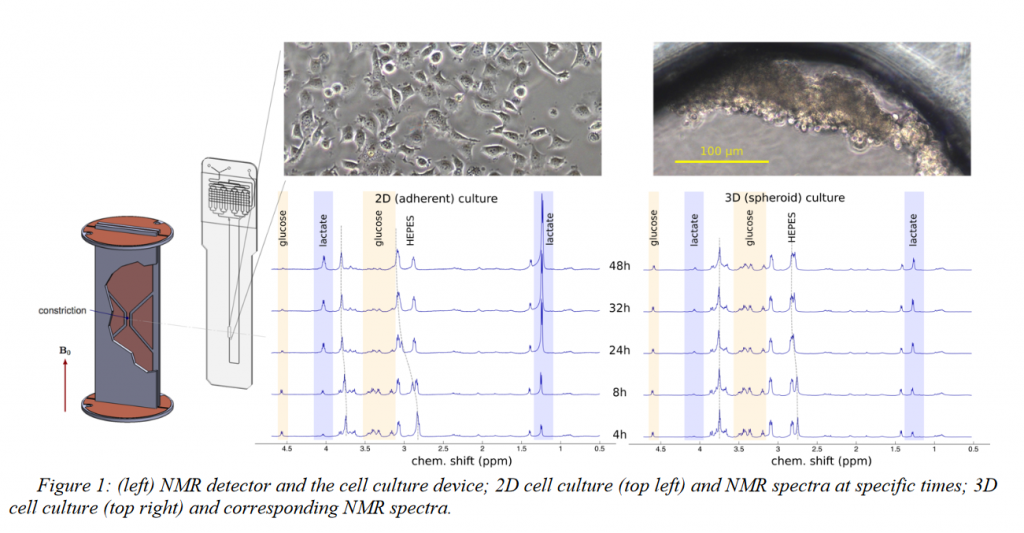
Figure 1 shows the 1HNMR spectra of 2D and 3D cultures up to 48 hours. 2D cells already started to produce more lactate compared to the 3D cell culture after 4 hours. At 48 hours 2D cell culture produces enough lactate to significantly shift the HEPES buffer resonances and consumed almost all the glucose. By contrast, 3D cells produced significantly less lactate. NMR spectra obtained in this way are of significant quality for unsupervised peak identification and metabolites present could be identified by mining the human metabolome database(figure 2). Figure 3 shows the viability analysis of 2D and Spheroid after 48 hours. The live stain (Green) indicates the healthy cells, whereas dead cells are stained as red. After 48 hours, 2D medium got enough acidified that cells are detaching, whereas cells survived strongly as spheroid.
ACKNOWLEDGEMENTS
This research project is funded by the horizon 2020 Framework program of the European Union.
REFERENCES
[1] F. Pampaloni, E. G. Reynaud, E.H.K. Stelzer, “The third dimension bridges the gap between cell culture and live tissue”. Nat Rev Mol Cell Biol. 8, 839, 2007
[2] G. Finch, A. Yilmaz, M. Utz, “An optimized detector for in-situ high-resolution NMR in microfluidic devices”. J. Mag. Res.262, 73-80, 2016
[3] D.S. Wishart, Y.D. Feunang, A. Marcu, A.C. Guo K. Liang, “ HMDB 4.0-The human metabolome database for 2018”. Nucleic Acids.Res.Jan 4; 4 6(D1): D608-17, 2018